A particular dream experiment is to directly watch atomic motions during a chemical event, such as a bond breaking. With the advent of ultrabright electron and x-ray sources, this has become possible.
Techniques such as coherent imaging allow us to directly watch atomic motions in complex systems, such as in a biological reaction or collective dynamics in a condensed matter system. In this research area we bring together multidisciplinary expertise in laser science, structural biology, chemistry, molecular physics, and imaging science and focus on the basic underlying concepts of systems from small molecules to amino acids and to protein complexes.
Research Focus B.1: Structure-function Correlation in Biological Systems
The research focus consists of three integrated research projects:
- B1.1: Atomically Resolved Reaction Dynamics of Model Systems
- B1.2: Atomic Perspective of Biological Functions
- B1.3: Higher Organizational Length Scales.
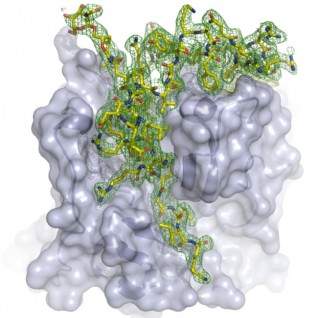
The LCLS structure reveals the natural inhibitor that binds in the enzyme’s active site
Nature as we perceive it from a scientific perspective is composed of fundamental building blocks at different levels of complexity – from atoms and molecules to complex and self-organizing structures.
In area B.1, we aim to understand and quantify the structure-function correlation in biological systems by investigating small, well defined and well controlled model systems. We start from isolated small molecules in the gas phase and proceed to amino acids and large biological macromolecular systems such as proteins and and protein complexes.
One goal is to determine the role and influence of dynamics on structure-function correlation at the single protein level. A direct observation of transition state processes is one of the holy grails of physical chemistry and can unify chemistry and biology at this molecular level. Another goal is to directly watch fundamental biological processes in living cells in nanometer and nanosecond resolution. For achieving our goals, we are using next-generation ultrafast and ultrabright XFEL and electron sources as well as advanced molecular spectroscopy techniques and novel instrumentation for sample preparation and handling.
Research Focus B.2 Expanding the Structural Basis of Biology with Novel Radiation Sources
The structural biology methods in Hamburg allow the analysis of three-dimensional atomic structures of a broad variety of biological macromolecules, ranging from single domain proteins to challenging multi-subunit protein complexes and molecular machines, including membrane proteins.
For example one research focus of the CUI research collaboration is the detailed analysis of a central syringe-like nanomachine, present in numerous bacterial pathogens is the type-three secretion System (TTSS). These macromolecular machines are capable to translocate an arsenal of specific bacterial toxins (effector proteins), some of which modulate host cell mechanisms for the benefit of the pathogen. Assembly, penetration of host cell membranes, and protein transport activity of translocons is far from being understood and methods that allow visualizing molecular dynamics of these assemblies in action represent a quantum leap in infection biology.
Further, time-resolved structure determination at hundreds of time points, to follow photo-stimulated ATP/GTP-driven reactions over a broad range of timescales are performed applying the new and upcoming radiation sources available at DESY. This opens up general methods to study most molecular machines in action.
A challenge is also represented by proteins or protein complexes currently not accessible due to limitations of the current standard methods. So far only about 15% of soluble proteins are found to form crystals large enough for protein crystallography and the situation is much worse for membrane-associated proteins. Particular membrane proteins play a crucial role in delivering small drug molecules across the cell membrane, and hence have an overwhelming impact on human health. Over 30% of human genes code for membrane proteins and 60% of all drugs target these proteins.
The novel ultra-intense X-ray and electron radiation sources provide entirely new opportunities to obtain new insights about structure-function-relationships in structural biology, beyond the recently believed feasibility limits in terms of molecular size, complexity, and structural dynamics of biological samples.
Research Focus B.3: New Methods for Atomically Resolved Imaging
Within this research topic the interdisciplinary and complementary expertise of CUI scientists in theoretical and experimental disciplines, including informatics, scattering physics, structural and systems biology, sample handling methods, molecular dynamics as well as laser, X-ray and electron source development are combined to investigate molecular structure and dynamics beyond current limitations.
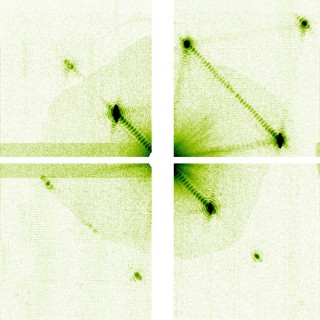
Detail of a single-pulse nanocrystal diffraction pattern showing fringes due to the crystal’s finite shape and beam coherence.
The unique radiation sources and expertise in instrumentation and detector development of this cluster of excellence provide exceptional opportunities to gain insights into the structure and dynamics of biological molecules and molecular assemblies. Chemical and biological processes are analyzed in atomic detail and observed up to the femtosecond time scale. The research collaboration investigates kinetic processes, molecular recognition events, functional and conformational domain movements as well as steps of protein folding, refolding, and aggregation, utilizing the FLASH soft X-ray FEL, the pulsed electron source REGAE, the high brilliance synchrotron radiation at PETRA III and the future European XFEL. The unique combination of highly coherent, intense radiation sources and novel imaging methods certainly boost these investigations.
The new method of serial femto second crystallography allows us to exploit nanocrystals, which in combination with new methods to phase diffraction data pave the way towards smaller and smaller samples, eventually allowing the analysis of uncrystallized molecular ensembles. For this step ahead the methodology of coherent diffraction scattering are developed further, both in the areas of experimental techniques and theoretical modelling. CUI scientists also establish new and advanced methods to prepare and score 2D molecular areas and 3D nanocrystal suspensions, most suitable for X-ray and electron diffraction experiments. The approach of in vivo crystallization can produce billions of nanocrystals required for subsequent X-ray FEL or electron diffraction experiments. This method was successfully applied by CUI scientists to solve the first unknown macromolecular structure with an X-ray FEL. We are developing this into a semi-automatic crystallization platform combining advanced tools to precisely monitor crystal growth and optimize the preparation of crystalline particles, too small to be observed by light microscopes.
Participating Research Groups
- Prof. M. Aepfelbacher
- Prof. C. Betzel
- Prof. R. Blick
- Prof. C. Bressler
- Prof. H. Chapman
- Prof. G. Grübel
- Prof. F. Kärtner
- Prof. J. Küpper
- Prof. R.J.D. Miller
- Prof. C. Morrison
- Prof. A. Pearson
- Prof. A. Rentmeister
- Prof. R. Santra
- Dr. M. Schnell
- Prof. M. Thorwart
- Prof. H. Tidow
- Dr. T. Tschentscher
- Prof. E. Weckert
- Prof. H. Weller
- Dr. M. Wilmanns
- Prof. B. Ziaja-Motyka

