Bacterial pathogens have developed sophisticated mechanisms to evade the immune system and spread in the human host. A team led by CUI member Prof. Martin Aepfelbacher of University Medical Center Hamburg-Eppendorf in collaboration with groups around CUI Prof. Christian Betzel (Universität Hamburg) and Dmitri Svergun (European Molecular Biology Laboratory in Hamburg) has succeeded in analyzing an important mechanism of a bacterial infection strategy. The scientists report their findings in the journal PLOS Pathogens. The results could be important for immune therapy as well as an effective treatment of infections.
Pathogenic Yersinia, including Yersinia pestis, the causative agent of bubonic plague, use a rather effective infection strategy: They even succeed in injecting a protein called YopM into host cells. YopM enters the nucleus and directly elevates the transcription of anti-inflammatory cytokines which suppress the host immune system. “It has been unclear so far through which mechanisms YopM shuttles between the cytoplasm and the nucleus of infected cells,” says CUI scientist Markus Perbandt, a coauthor of the study. The team has just identified a nuclear protein which transports YopM out of the nucleus and thereby enables its nucleocytoplasmic shuttling. The researchers could demonstrate that the nuclear level of YopM is proportional to its immunosuppressive effect. Aepfelbacher: “Thus, the newly identified mechanism of nucleo-cytoplasmic shuttling of YopM directly contributes to the ability of the bacteria to cause infection.”
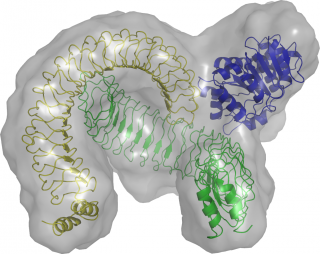
Structural model of the YopM/DDX3 complex based on small angle X-ray scattering and crystallographic data. © PLOS Pathogen
The protein enabling nuclear export of YopM was identified as the DEAD box RNA helicase DDX3. DDX3 itself is transported out of nucleus through the export factor CRM1. In collaboration with the groups of Christian Betzel und Dmitri Svergun YopM was crystallized and its structure was solved. On this basis the solution structure of the YopM:DDX3 complex could be recorded by small angle X-ray scattering. Structural analysis indicated that YopM forms a dimer and that the N-terminal domain (domain1) of DDX3 interacts with the interface created by the two YopM monomers. Moreover, in an earlier study CRM1 was described to interact with the C-terminus of DDX3 (domain 2).
Based on the new data, a model complex comprising CRM1 in addition to YopM and DDX3 could be assembled. “These discoveries could have diverse implications,” Aepfelbacher explains. “Knowing how YopM exactly operates, could help in using its immunosuppressive activity as a new therapeutic principle. Furthermore, inhibitors of YopM might be considered as novel antiinfective agents.”
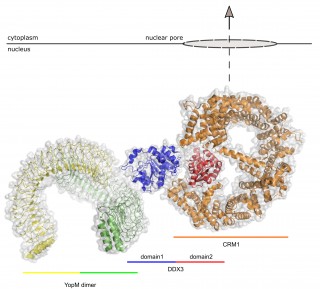
Potential structure of a nuclear export complex comprising YopM, DDX3 and CRM1. The hypothetical model of the nuclear export complex is based on the experimentally solved structures and the published interaction sites between these proteins. © Martin Aepfelbacher
Citation:
Berneking L, Schnapp M, Rumm A, Trasak C, Ruckdeschel K, Alawi M, Grundhoff A, Kikhney AG, Koch-Nolte F, Buck F, Perbandt M, Betzel C, Svergun DI, Hentschke M, Aepfelbacher M.
“Immunosuppressive Yersinia Effector YopM Binds DEAD Box Helicase DDX3 to Control Ribosomal S6 Kinase in the Nucleus of Host Cells”
PLoS Pathog. 2016 Jun 14;12(6):e1005660
DOI: 10.1371/journal.ppat.1005660

